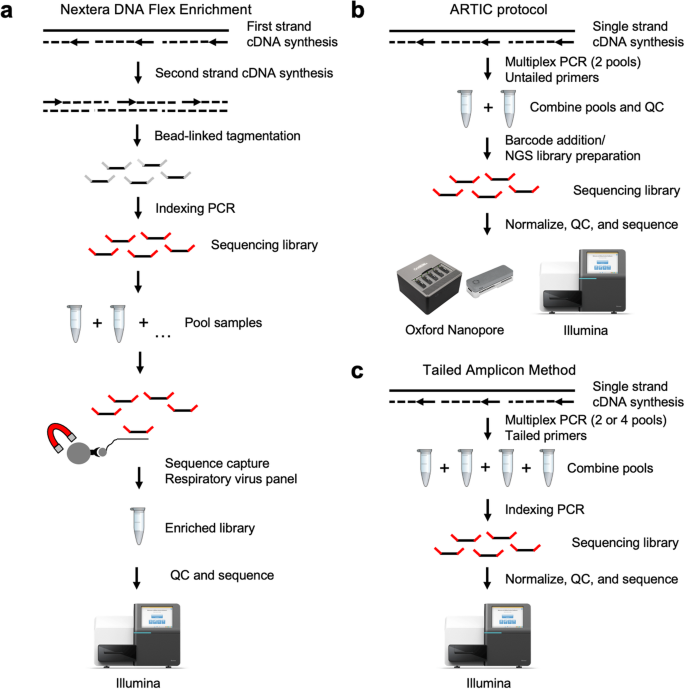
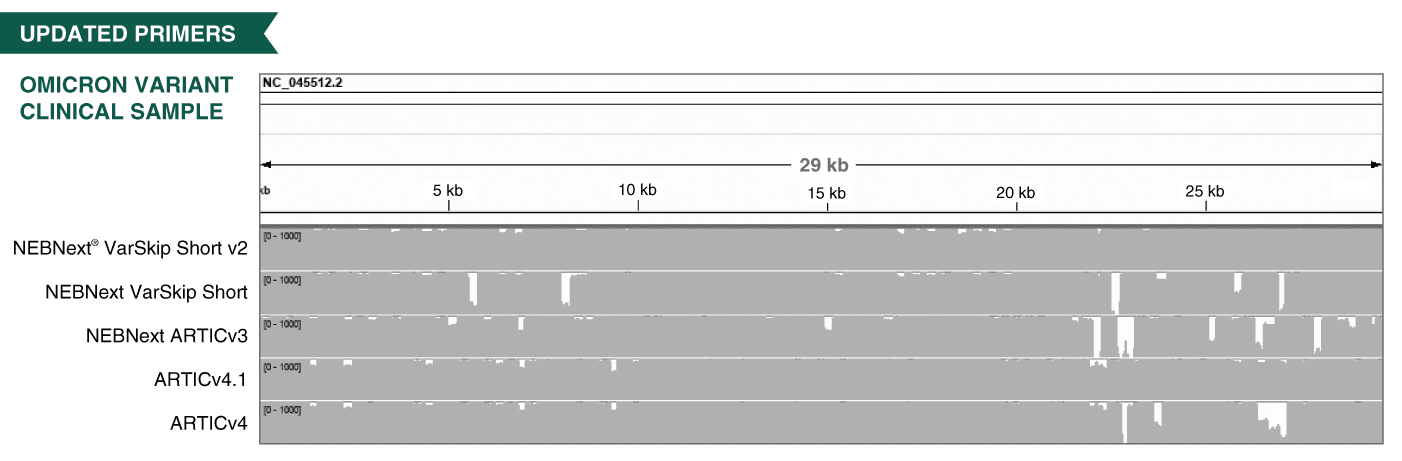
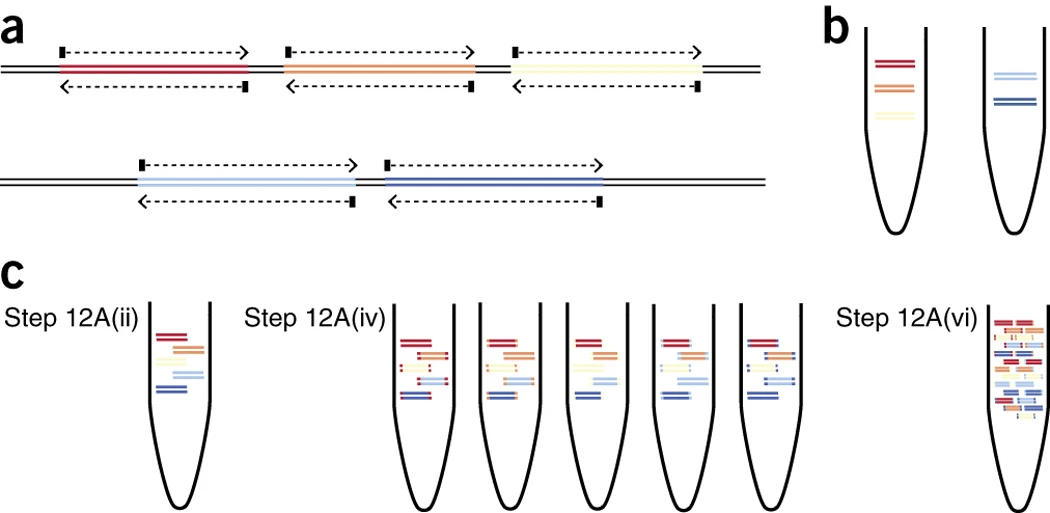
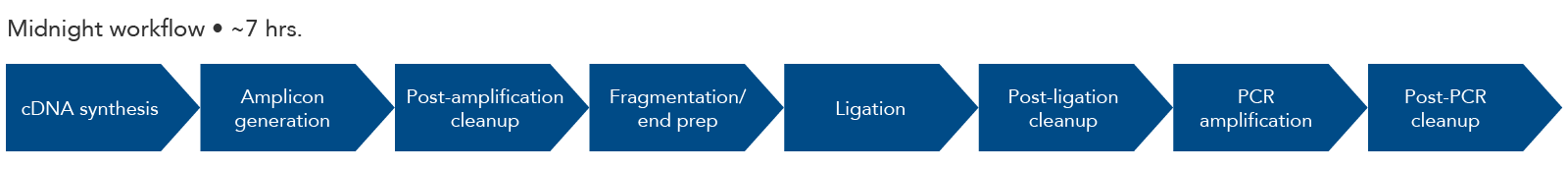
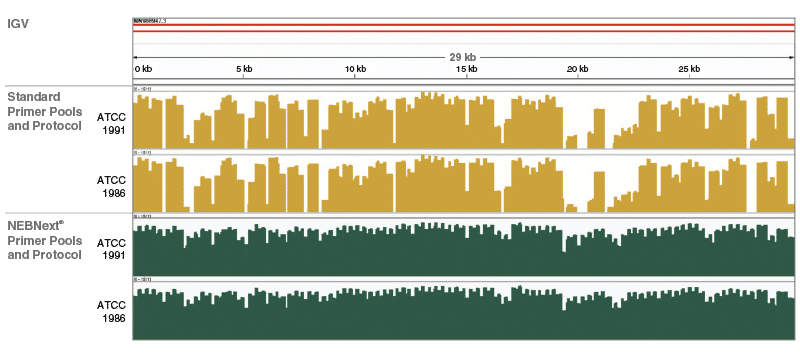
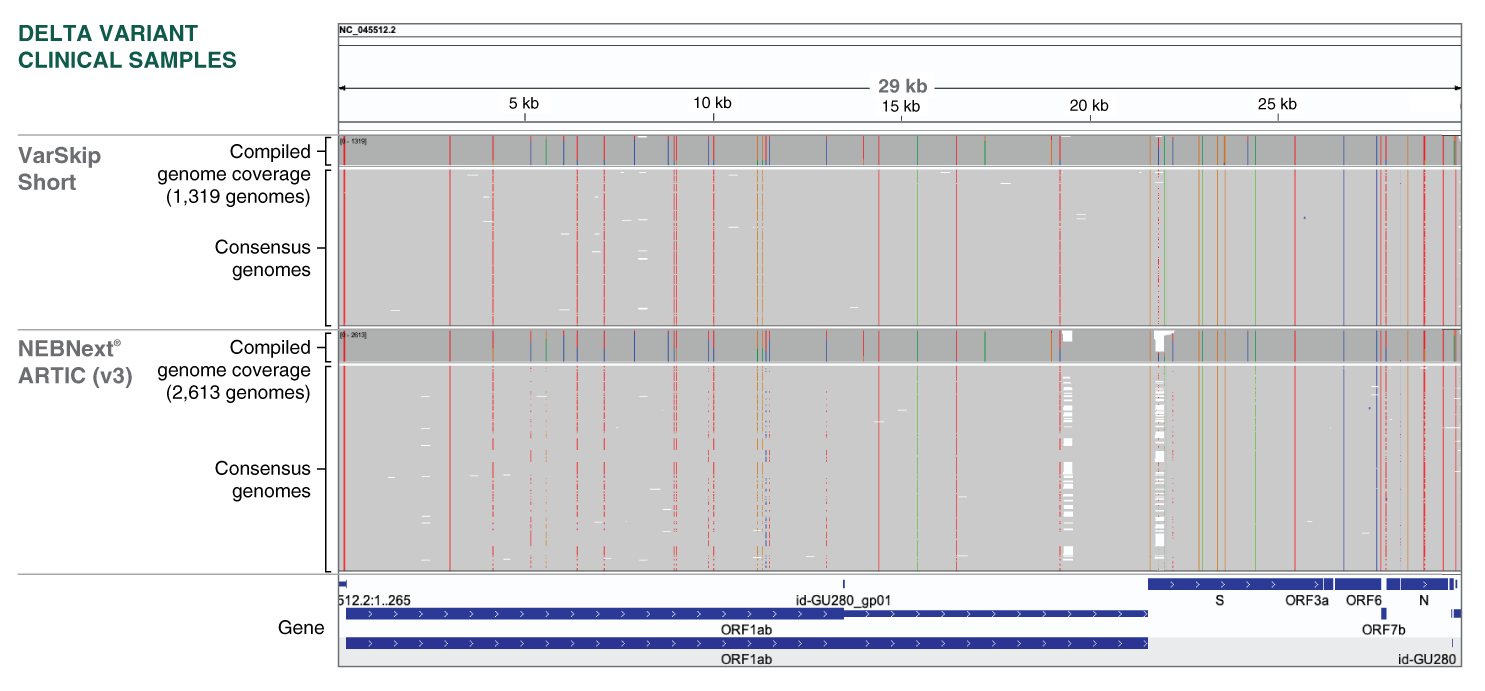
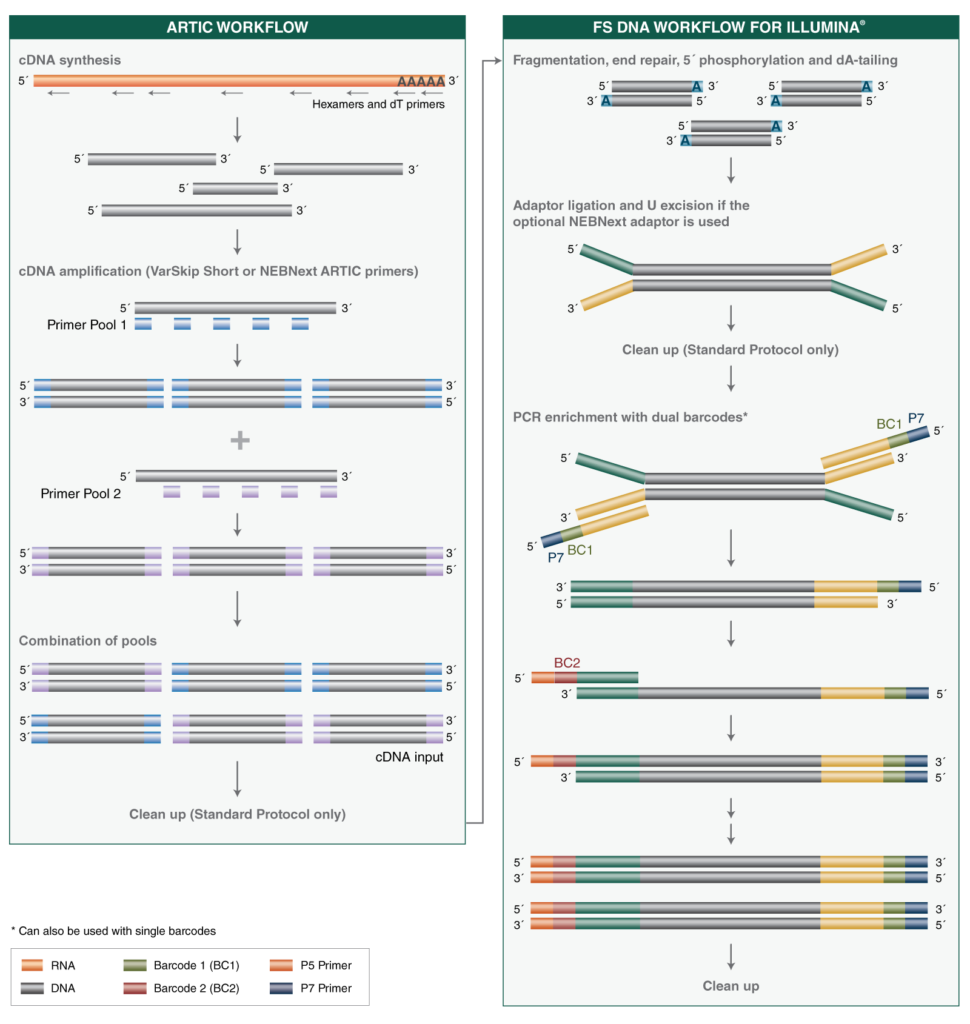
periscope: sub-genomic RNA identification in SARS-CoV-2 ARTIC Network Nanopore Sequencing Data | bioRxiv
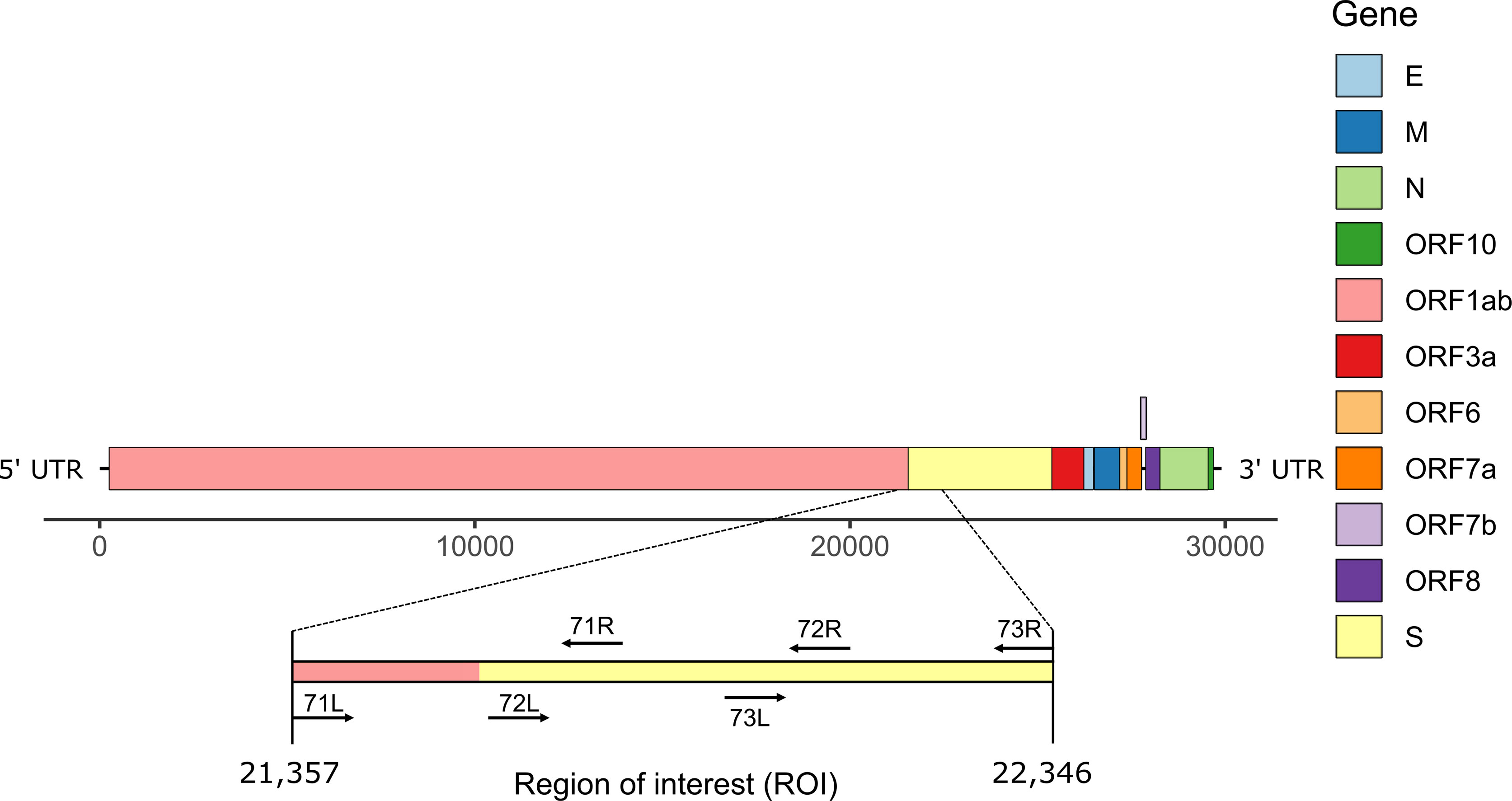
Frontiers | Investigating the Extent of Primer Dropout in SARS-CoV-2 Genome Sequences During the Early Circulation of Delta Variants

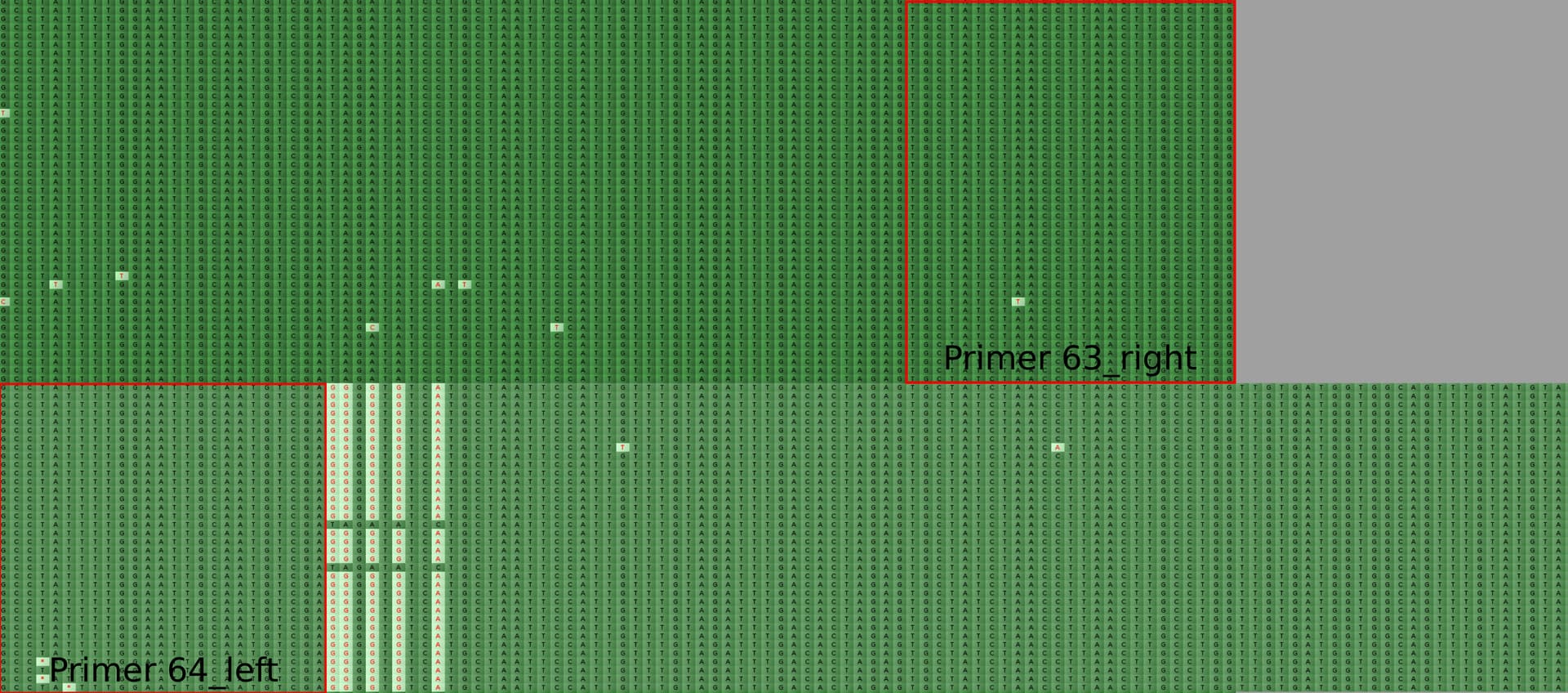
Missing G21987A mutation in SARS-CoV-2 delta variants due to non-specific amplification by ARTIC v3 primers - nCoV-2019 Genomic Epidemiology - Virological

Monitoring SARS-CoV-2 variants alterations in Nice neighborhoods by wastewater nanopore sequencing - The Lancet Regional Health – Europe
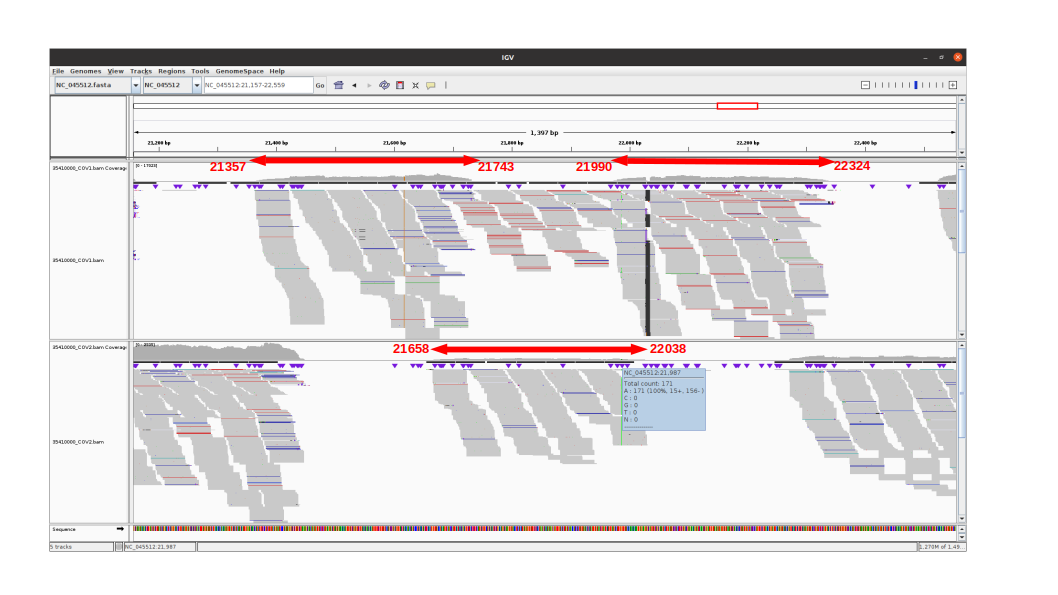
Missing G21987A mutation in SARS-CoV-2 delta variants due to non-specific amplification by ARTIC v3 primers - nCoV-2019 Genomic Epidemiology - Virological
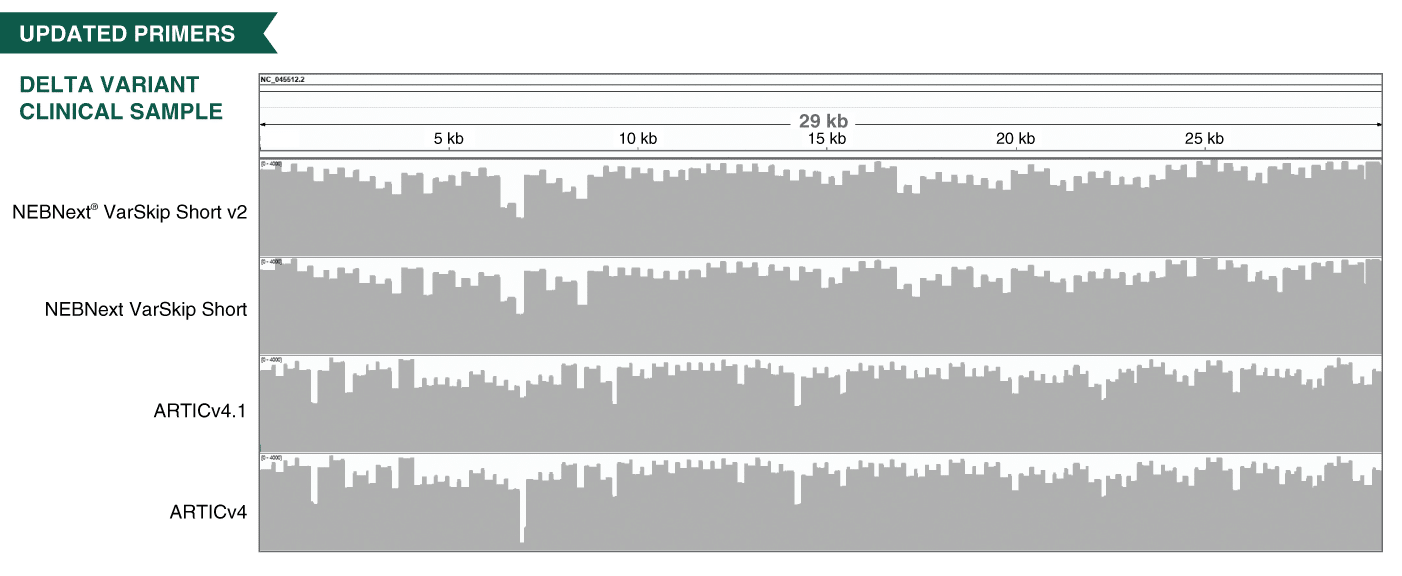
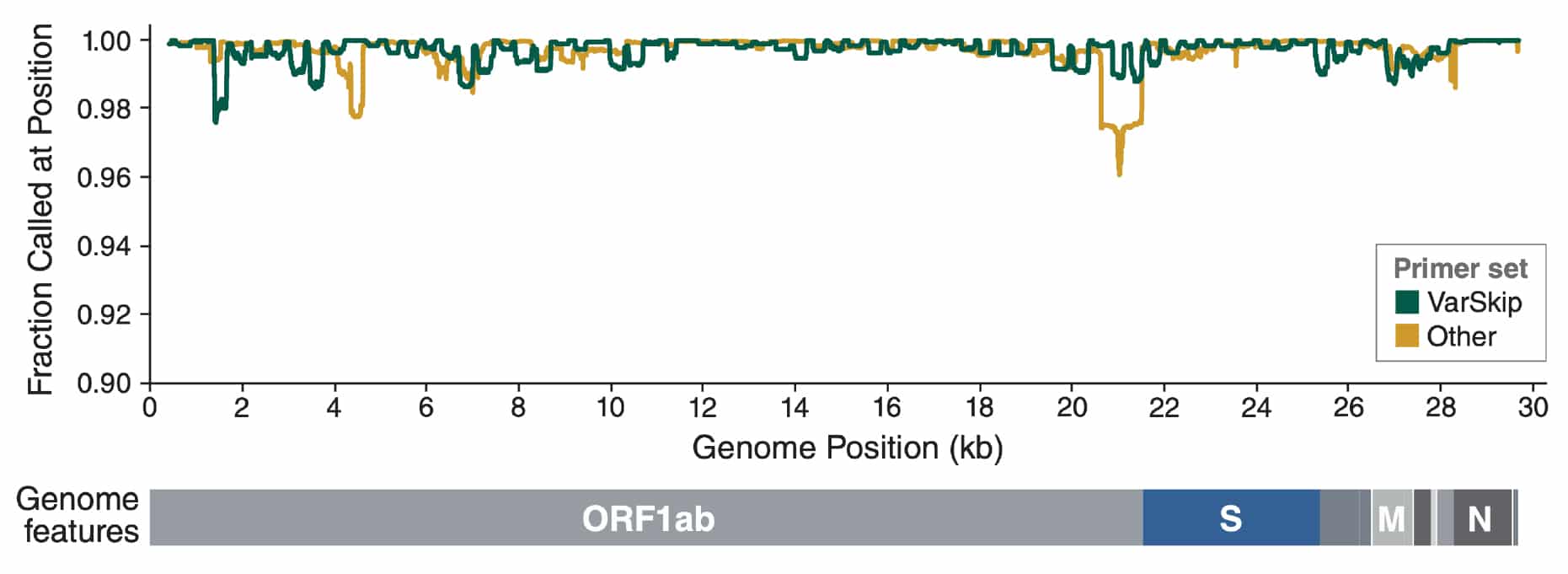
A proposal of alternative primers for the ARTIC Network's multiplex PCR to improve coverage of SARS-CoV-2 genome sequencing

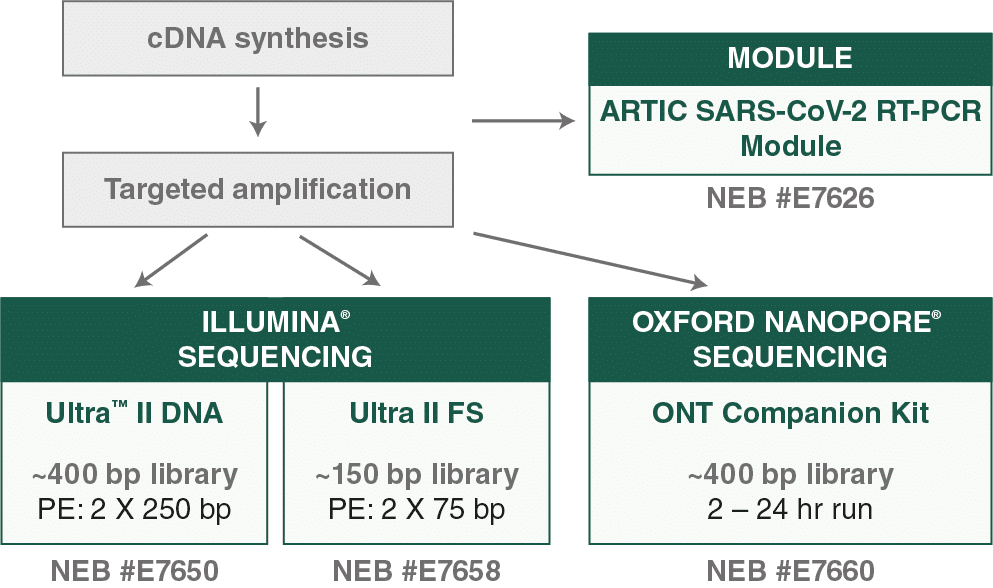
ARTIC Network provides protocol for rapid, accurate sequencing of novel coronavirus (nCoV-2019): first genomes released